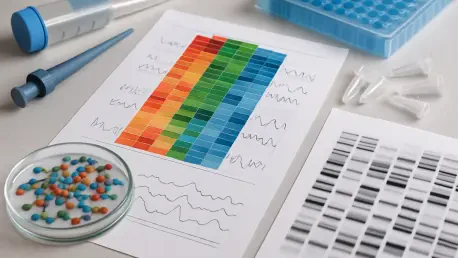
Imagine a world where scientists can peek into the intricate dance of cells within a tissue, mapping out exactly where and how genes are expressed, revealing secrets of diseases like cancer or the mysteries of brain development. Spatial transcriptomics offers this remarkable capability, allowing researchers to visualize gene activity while preserving the spatial context of cells in their natural environment. This technology has the potential to redefine biological research, offering insights that traditional methods simply can’t capture. Yet, despite its promise, the field faces a glaring problem: inconsistency. Variability in methods, data quality, and reproducibility across labs has created a bottleneck, slowing down the pace of discovery and casting doubt on results.
The Spatial Touchstone project, spearheaded by St. Jude Children’s Research Hospital in Memphis, Tennessee, steps into this gap as a game-changer. Led by Jasmine Plummer, PhD, director of the St. Jude Center for Spatial Omics, this multi-institutional initiative unites global expertise to tackle the field’s toughest challenges. Highlighted in a recent issue of Nature Biotechnology, the project aims to bring standardization and reliability to spatial transcriptomics, ensuring that this cutting-edge technology lives up to its transformative potential. It’s not just about solving a technical puzzle; it’s about laying a foundation for future breakthroughs that could impact everything from cancer treatments to neurological research.
Tackling the Core Challenges in Spatial Transcriptomics
Confronting Variability in Research Practices
Spatial transcriptomics holds immense promise, but its rapid growth has outpaced the development of consistent practices, leading to a frustrating lack of uniformity. Researchers often find themselves comparing apples to oranges when analyzing data across labs due to differences in sample preparation, imaging platforms, or processing techniques. This variability muddies the waters, making it nearly impossible to discern whether observed differences stem from genuine biological insights or mere technical artifacts. The Spatial Touchstone project dives headfirst into this issue, aiming to establish clear benchmarks that can guide the field. By creating a unified framework, it seeks to eliminate the guesswork, ensuring that scientists can trust their data reflects true biology rather than procedural hiccups.
Moreover, the absence of established quality metrics has long been a thorn in the side of spatial transcriptomics research. Without a baseline to measure against, labs risk wasting resources on datasets that fail to meet scientific standards. The frustration is palpable—time and funding are precious, and flawed experiments can set progress back significantly. Spatial Touchstone addresses this by introducing tools like the Spatial QM software, which provides tailored performance indicators for various tissues and platforms. This isn’t just a Band-Aid solution; it’s a proactive step to empower researchers with the means to validate their work early in the process, saving effort and building confidence in their findings.
Building a Foundation for Trustworthy Data
Beyond addressing immediate variability, the project focuses on the broader need for trust in scientific output. Spatial transcriptomics experiments are resource-intensive, often requiring substantial investments of time, money, and expertise. When results can’t be replicated due to inconsistent methods, the credibility of the entire field suffers. Spatial Touchstone pushes for a cultural shift toward rigorous standards, drawing inspiration from fields like genomics where standardization has paved the way for monumental discoveries. By setting clear expectations, it aims to create an environment where researchers can build on each other’s work without second-guessing the data.
Additionally, the initiative emphasizes the importance of tackling errors at their source. Issues like poor sample handling or mismatched equipment settings can skew results before analysis even begins. Through comprehensive guidelines and reference datasets covering a range of tissue types, Spatial Touchstone offers a roadmap for identifying and correcting such problems upfront. This preventative approach minimizes the risk of downstream complications, ensuring that every step of the research process aligns with a shared vision of quality. It’s about creating a solid bedrock on which the future of spatial transcriptomics can stand firm.
Harnessing Collaboration and Innovation for Global Impact
Uniting Expertise Across Borders
One of the standout strengths of Spatial Touchstone lies in its collaborative spirit, bringing together leading minds from institutions like St. Jude, Weill Cornell Medicine, and the University of Adelaide. This isn’t a solo endeavor but a global coalition, uniting researchers such as Christopher Mason and Luciano Martelotto to pool their expertise. By integrating diverse perspectives, the project crafts solutions that resonate across different research contexts. The inclusion of datasets from six tissue types—ranging from breast to pancreas, and covering both healthy and diseased states—ensures that the resources developed are not narrowly focused but widely applicable, addressing the needs of various studies from cancer to neurobiology.
Equally important is how this collaboration transcends geographical boundaries to foster a shared mission. The project doesn’t just compile data; it weaves together a network of knowledge that amplifies impact. Publicly available datasets are combined with newly curated ones, creating a rich repository that serves as a global reference. This approach breaks down silos, encouraging labs worldwide to contribute to and benefit from a collective pool of resources. The result is a unified front against the challenges of spatial transcriptomics, proving that when the scientific community works as one, the potential for progress multiplies exponentially.
Democratizing Access Through Open Science
Accessibility forms another cornerstone of Spatial Touchstone’s vision, ensuring that cutting-edge research isn’t confined to a privileged few. By making datasets, software, and protocols freely available online, the project levels the playing field for smaller labs or those with limited funding. In a field where high-end equipment and substantial budgets often dictate who can participate, this open science ethos is a breath of fresh air. It invites contributions from a broader range of scientists, enriching the field with diverse insights and accelerating collective discovery.
Furthermore, this transparency builds trust and fosters a culture of shared progress. When resources like the Spatial Touchstone Portal (STP) or the Spatial QM software are accessible to all, it reduces redundant efforts and encourages collaboration over competition. Labs no longer need to reinvent the wheel; they can tap into established tools to enhance their work. This democratization doesn’t just benefit individual researchers—it strengthens the entire ecosystem of spatial transcriptomics, aligning with broader scientific trends toward openness and inclusivity that promise to drive innovation forward.
Empowering Researchers with Cutting-Edge Tools
Streamlining Quality Control with Practical Solutions
A defining feature of Spatial Touchstone is its suite of user-friendly tools designed to bring precision to spatial transcriptomics research. The Spatial Touchstone Portal (STP) stands out as a vital resource, allowing scientists to screen samples against established benchmarks before committing to full-scale analysis. This early validation step is a game-changer, catching potential issues like substandard data quality before they derail an entire project. Instead of sinking time into flawed experiments, researchers can pivot quickly, focusing their efforts on reliable datasets. It’s a practical solution that speaks directly to the real-world frustrations of working in a high-stakes field.
In addition, the Spatial Touchstone Standard Operating Procedures (STSOP) provide a detailed blueprint for consistent experimentation. From tissue preparation to data acquisition, these guidelines ensure that every stage of the process adheres to a high standard. This isn’t about stifling creativity but rather about creating a common language for labs worldwide. By reducing variability in methods, STSOP helps guarantee that results can be compared and replicated across different settings, fostering a sense of reliability that’s been elusive in spatial transcriptomics until now. It’s a tool that balances structure with flexibility, meeting researchers where they are.
Enhancing Performance with Targeted Metrics
Complementing these resources is the Spatial QM software, which offers a sophisticated way to track performance across diverse platforms and tissue types. Recognizing that not all labs use the same equipment or study the same biological systems, this tool provides tailored metrics to evaluate data quality. Whether it’s assessing imaging accuracy or processing efficiency, Spatial QM ensures that researchers have a clear picture of where their experiments stand. This specificity is crucial in a field where one-size-fits-all solutions often fall short, and it empowers scientists to make informed decisions about their next steps.
Beyond mere measurement, Spatial QM serves as a diagnostic tool, helping pinpoint where errors might creep in. If a dataset underperforms, the software can highlight whether the issue lies in sample handling, platform settings, or another variable. This targeted feedback loop saves not just time but also the intellectual energy that might otherwise be spent troubleshooting blindly. By integrating seamlessly into existing workflows, it minimizes disruption while maximizing impact, proving that innovation in spatial transcriptomics doesn’t have to be complicated to be effective. It’s a testament to Spatial Touchstone’s commitment to practical, actionable solutions.
Paving the Way for a Reliable Future in Transcriptomics
Balancing Innovation with Methodical Rigor
Spatial transcriptomics has surged forward with breathtaking speed, driven by the thrill of uncovering cellular interactions in unprecedented detail. However, this rapid expansion has sometimes come at the cost of careful methodology, leaving gaps in reliability that could undermine long-term progress. Spatial Touchstone steps in to strike a critical balance, advocating for a slower, more deliberate approach to ensure that the field’s foundation is rock-solid. It’s not about dampening excitement but channeling it into sustainable growth. By prioritizing rigor over rush, the project ensures that each new discovery builds on a bedrock of trustworthy data, setting a standard for how emerging technologies should evolve.
Looking back, the emphasis on standardized practices through comprehensive datasets and tools like STSOP proved to be a turning point. The field had grappled with inconsistencies for too long, and Spatial Touchstone’s methodical framework offered a way to navigate those challenges with confidence. The lesson was clear: innovation thrives when paired with discipline. As the scientific community moves forward, this initiative’s influence serves as a reminder that taking the time to get the basics right can unlock exponential potential down the line.
Setting a Precedent for Emerging Technologies
Reflecting on the journey, Spatial Touchstone not only addressed immediate hurdles in spatial transcriptomics but also carved out a blueprint for other cutting-edge fields. The collaborative model, blending global expertise with open-access resources, demonstrated how to tackle systemic issues in science collectively. It showed that standardization doesn’t stifle progress—it amplifies it by creating a shared starting point. For emerging technologies facing similar growing pains, this project laid down a path worth following, proving that investment in quality control can yield dividends across disciplines.
As the field continues to evolve, the next steps involve expanding these resources to cover even more tissue types and platforms, ensuring that no corner of spatial transcriptomics is left behind. There’s also a push to integrate advanced analytics, like machine learning, to refine quality metrics further. These future considerations keep the momentum alive, encouraging researchers to build on Spatial Touchstone’s legacy. It’s a call to action for the scientific community to keep refining, sharing, and innovating, ensuring that the promise of mapping gene expression in space transforms from a vision into a lasting reality.